Table of Contents
Introduction: The Importance of Protein Stability
Understanding protein stability is fundamental for modern biochemistry, pharmaceutical research, and biotechnology. Protein stability determines not only how long a therapeutic protein can remain active in solution but also its suitability for formulation, storage, and transport (Wang 1999). From enzyme engineering to drug development and structural biology studies, the ability to accurately assess and optimize protein stability enables researchers and innovators to advance life science applications while minimizing risks due to aggregation, denaturation, or loss of activity.
Recent publications underscore the role of protein stability in drug formulation and bioprocessing—where even small changes in buffer composition or temperature can profoundly affect protein fold and therapeutic outcomes (Durowoju et al. 2017).
Factors Influencing Protein Stability
Protein function is critically dependent on the integrity of its three-dimensional structure—the native conformation. This structure can be perturbed by a variety of physicochemical factors:
- Temperature: Elevated temperatures may unfold proteins, causing irreversible loss of function or aggregation.
- pH: Changes in pH disrupt ionic interactions and hydrogen bonds, impacting structural stability.
- Ionic strength: Salts can stabilize or destabilize proteins depending on specific interactions with charged side chains.
- Ligand binding: Small molecules, cofactors, or drugs that bind to proteins may stabilize or destabilize specific conformations (Durowoju et al. 2017).
- Genetic mutations: Single point mutations may enhance or reduce stability, influencing protein behavior under physiological conditions.
Systematic studies reveal that adjusting buffer chemistry and controlling temperature during handling and storage are among the most effective interventions to preserve protein activity (Wang 1999).
Fundamentals: What Does Protein Stability Mean?
Protein stability refers to the resilience of a protein’s native structure under various conditions. It encompasses:
- Thermodynamic stability: The tendency of a protein to remain folded rather than denature.
- Kinetic stability: The rate at which a protein transitions to an unfolded or aggregated state.
- Reversible changes: Some folding transitions can be reverted by restoring benign conditions (e.g., refolding after mild denaturation).
- Irreversible changes: Severe processing (e.g., high temperatures or chemical agents) may lead to aggregation or unfolding that cannot be reversed.
Stability is often quantified by the melting temperature (Tm), at which half of the protein population is denatured—a key parameter measured in stability assays (Durowoju et al. 2017).
Methods to Assess Protein Stability
Multiple orthogonal techniques have been developed to study biomolecular stability and interactions. Selecting the appropriate method depends on the research question, protein type, and available instrumentation.
Differential Scanning Calorimetry (DSC)
DSC is an essential tool for precise stability studies in proteins (Durowoju et al. 2017). By measuring the heat required to unfold a protein sample as temperature increases, DSC determines:
- Denaturation temperature (Tm): Indicates thermal stability.
- Enthalpy changes (ΔH): Reveals folding energy transitions.
- Unfolding profiles: Characterize reversible or irreversible transitions.
DSC is valued for its accuracy in detecting subtle transitions—even for proteins at low concentrations—and is especially powerful for characterizing variants, thermal formulation behavior, and ligand-induced stability shifts (Qualification of a DSC Method 2020) .
Other Analytical Methods
Dynamic Light Scattering (DLS): Measures hydrodynamic radius and detects protein aggregation, complementing thermal stability assays (Mirasol et al. 2021).
UV-Vis Spectroscopy: Monitors unfolding by tracking changes in absorbance related to aromatic side chains or chromophores.
Isothermal Titration Calorimetry (ITC): Quantifies binding interactions between proteins and ligands; useful for detecting stability changes upon ligand association (Durowoju et al. 2017).
Circular Dichroism (CD) Spectroscopy: Probes secondary structure changes during folding or denaturation (e.g., alpha-helix to beta-sheet transitions).
Studying Interactions Affecting Stability
Proteins continually interact with their molecular environment. Assessing how these interactions affect stability is essential for research and development:
- Ligand binding: DSC and ITC are used to determine binding affinities and the stabilizing or destabilizing effects of small molecules, drugs, and cofactors (Durowoju et al. 2017)
- Protein aggregation: DLS identifies changes in aggregation propensity during heat or stress exposure (Wang 1999).
- pH dependence: By conducting DSC runs at different pH values, researchers gain insights into the role of protonation states in folding.
- Genetic variants: Comparing Tm or aggregation profiles via DSC and DLS allows evaluation of stability conferred by point mutations or engineered sequence changes (Mirasol et al. 2021).
How Thermal Methods Illuminate Molecular Interactions
Thermal techniques like DSC not only yield Tm and enthalpy—but when combined with ligand or buffer titration protocols, provide deep insights into binding affinities and structural stabilization (Durowoju et al. 2017). For example, a ligand that increases the Tm of a protein suggests direct stabilizing interaction relevant to drug design or formulation.
Practical Insights and Measurement Parameters
Interpreting protein stability data requires attention to experimental conditions and typical outputs:
- Heating rate: Influences kinetic vs. thermodynamic stability detection; slower rates favor equilibrium measurements.
- Sample concentration: Sufficient concentration ensures reliable signal without aggregation interference.
- Buffer composition: Compare stabilizing effects of different buffers and excipients on Tm and unfolding enthalpy (Wang 1999).
- Control samples: Always compare to well-defined standards—such as wild type vs. mutant proteins or buffer-only controls.
Application
Example Applications
- Comparing stability profiles of monoclonal antibodies for formulation development (Qualification of a DSC Method 2020).
- Evaluating enzyme variants for industrial biocatalysis requiring thermal resilience.
- Screening protein-ligand binding affinities in early-stage drug discovery and structural biology (Durowoju et al. 2017).
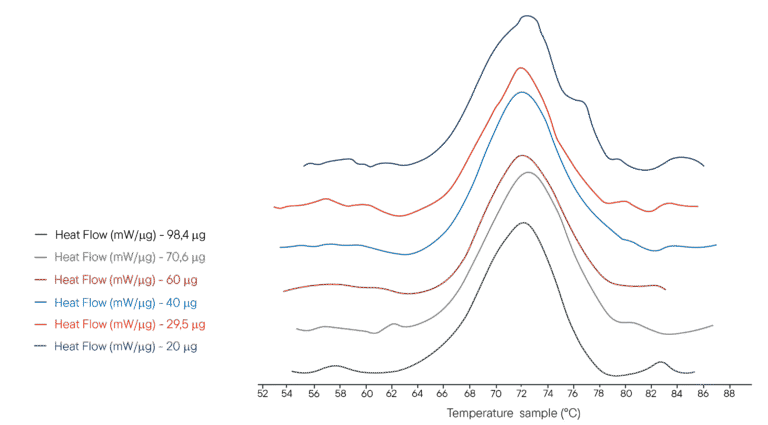
Application example: Thermal denaturation of Lysozyme
100µL of several solutions of lysozyme in PHB buffer were placed in a 100µL crucible. Different concentrations were used, representing different masses of lysozyme analysed respectively. The reference crucible was filled with the same volume of PHB buffer. The Ultimate DSC was programmed to perform a temperature ramp from 40°C to 95°C at 1°C/min.
The thermograms obtained are shown on the right.The Ultimate DSC allows the enthalpy and denaturation temperature of Lysozyme to be measured using 5 to 10 less sample than currently available instruments. Each thermogram was obtained in less than 60 minutes, allowing rapid analysis. The Ultimate DSC saves large amounts of sample and allows you to work faster.
Selecting the Right Method for Your Research
Choosing the optimal analytical method depends on your specific protein, research goal, and available resources:
| Method | Key Insights | Typical Applications |
|---|---|---|
| DSC | Tm, ΔH, unfolding profile | Formulation, variant screening, stability assays |
| DLS | Aggregation, size distribution | Formulation, quality control, aggregation analysis |
| ITC | Binding affinity, thermodynamics | Drug design, ligand screening, protein-protein interactions |
| CD-Spectroscopy | Secondary structure change | Conformational studies, denaturation monitoring |
| UV/Vis Spectroscopy | Folding/unfolding, aggregation | Quick screening, routine assay |
For detailed comparison, recent reviews highlight how integrating multiple techniques, including precise calorimetric and spectroscopic measurements, leads to robust assessment of protein stability and suitability for downstream applications (Mirasol et al. 2021; Durowoju et al. 2017).
Conclusion & Next Steps
Understanding and controlling protein stability is central to successful biomolecules research, pharmaceutical formulation, and drug development. Thermal analysis, particularly using DSC, provides robust, quantifiable insights into protein behavior under practical conditions, helping you to make data-driven decisions with confidence.
If you are seeking to advance your protein stability investigations, the Linseis UDSC L64 provides state-of-the-art calorimetric capabilities tailored for biomolecular analysis and formulation development. Discover more about the UDSC L64’s performance and versatile application spectrum on our calorimetry solutions page, or reach out to our specialized team for individualized consultation regarding your research needs.
Selected Literature for Further Reading
- Durowoju, I.B., Bhandal, K.S., Hu, J., Carpick, B. and Kirkitadze, M. (2017) Differential Scanning Calorimetry—A Method for Assessing the Thermal Stability of Proteins, Journal of Visualized Experiments, (121), e55262. https://pmc.ncbi.nlm.nih.gov/articles/PMC5409303/
- Wang, W. (1999) ‘Instability, stabilization, and formulation of liquid protein pharmaceuticals’, International Journal of Pharmaceutics, 185(2), pp. 129-188. https://doi.org/10.1016/S0378-5173(99)00152-0
- Mirasol, F., Wypych, J. and Kopec, B. (2021) Stability Testing of Protein Therapeutics Using DLS, Pharmaceutical Technology. https://www.pharmtech.com/view/stability-testing-of-protein-therapeutics-using-dls
- Qualification of a Differential Scanning Calorimetry Method for Biophysical Characterization of Monoclonal Antibodies (Open Access). https://www.openaccessjournals.com/articles/qualification-of-a-differential-scanning-calorimetry-method-for-biophysical-characterization-of-monoclonal-antibodies-an.pdf